Disclaimer: The views expressed in this blog are meant to provide a summary and overview of the COVID variants. They are not meant to replace official communication, warnings, or guidelines regarding the variants. For the most up-to-date information, please refer to the Ministry of Health and Welfare’s Official COVID-19 website (https://www.mohfw.gov.in/).
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), commonly known as COVID-19, is a type of virus. A virus is known to mutate due to environmental changes and SARS-CoV-2 is no different. Since its first discovery in Wuhan on 7tth January 2020, by the Chinese authorities, there have been various changes in the genetic code of the virus. At the time of uploading this article, 14 variants of the virus are already in circulation among the human population. There are two ways of designating a virus: as a variant of concern (VOC) or as a variant under investigation (VUI). A VOC is a variant in which the mutation is beneficial to the virus as it makes it more transmissible and as a result becomes a cause of concern. On the other hand, a VUI is a variant that meets enough criteria to be noticed but requires further observation before being placed on the VOC list. The criteria used for these designations kept in mind are:
1. Increased transmissibility
2. Increased morbidity
3. Increased mortality
4. Increased risk of “long COVID”
5. Ability to evade detection by diagnostic tests
6. Decreased susceptibility to antiviral drugs (if and when such drugs are available)
7. Decreased susceptibility to neutralizing antibodies, either therapeutic (e.g., convalescent plasma or monoclonal antibodies) or in laboratory experiments
8. Ability to evade natural immunity (e.g., causing reinfections)
9. Ability to infect vaccinated individuals
10. Increased risk of particular conditions such as multisystem inflammatory syndrome
11. Increased affinity for particular demographic or clinical groups, such as children or immunocompromised individuals.
The table below displays the various variants, where and when they were first discovered along with a connection with relevant information pertaining to the virus.
| S.No | Name | Where | When | Relevant Information |
| 1 | Lineage B.1.1.7 | Kent, United Kingdom | December, 2020 | Detected in over 120 countries. This variant increased transmissibility by 40 to 80%. Potential to increase severity. |
| 2 | Lineage B.1.1.207 | Nigeria | August, 2020 | Detected in 10 countries. Has a shared mutation (P681H) with the B.1.1.7. |
| 3 | Lineage B.1.1.317 | Russia | December, 2020 | Contains the N501Y mutation that is also present in B.1.351. Strain is known to evade neutralising antibodies. |
| 4 | Lineage B.1.1.318 | UK | February, 2021 | Following assessments, the variant was designated a VUI on 24 February. It contains the E484K mutation, which is also found in 2 existing VUIs present in the UK but does not feature the N501Y mutation, present in all variants of concern. |
| 5 | Lineage B.1.351 | South Africa | December, 2020 | Has 3 mutations the N501Y, K417N, and E484K. Increased transmission. This mutant has a weakened neutralizing effect by antibodies. Potential to increase severity. |
| 6 | Lineage B.1.429 | California, USA | July, 2020 | Only 3.2% of all cases reported in the USA were caused by this variant. While the variant displayed increased transmissibility, it was less transmissible than B.1.1.7 |
| 7 | Lineage B.1.525 | UK | February, 2021 | B.1.525 differs from all other variants as it has both the E484K-mutation and a new F888L mutation. This variant does not carry the N501Y mutation. Increased transmission. This mutant has a weakened neutralizing effect by antibodies. |
| 8 | Lineage B.1.526 | New York City, USA | November, 2020 | In a pattern mirroring the B.1.429 variant of California, B.1.526 was initially able to reach relatively high levels in some states. Increased transmission. This mutant has a weakened neutralizing effect by antibodies. |
| 9 | Lineage B.1.617 | India | October, 2020 | It has 2 sub-variants B.1.617.2 and B.1.617.3. The B.1.617.2 (which notably lacks mutation at E484Q) spreads more quickly than the parent version of the virus. Known increased transmission. |
| 10 | Lineage B.1.618 | India | October, 2021 | It has a mutation called E484K which is the same mutation as the South African variant. As of 23 April 2021, the CoV-Lineages database showed 135 sequences detected in India, with single-figure numbers in each of eight other countries worldwide. Has the D614G mutation that is known to increase infectivity. |
| 11 | Lineage B.1.620 | Lithuania | March, 2021 | As of now, scientists have found that this lineage contains an E484 mutation. |
| 12 | Lineage P.1 | Manaus, Brazil and Japan | January, 2021 | This variant has been designated a ‘Variant of Concern’ because it shares some important mutations with the variant (B1.351) first identified in South Africa, similarly to E484K and N501Y. Increased transmission. This mutant has a weakened neutralizing effect by antibodies. |
| 13 | Lineage P.2 | South Brazil | October, 2020 | The P.2 lineage, which was first described in October 2020, only harbours the E484K mutation. This lineage, which is considered a variant of interest (VOI) was the most prevalent variant detected in several Brazilian states during late 2020 and early 2021. Increased transmission. This mutant has a weakened neutralizing effect by antibodies. |
| 14 | Lineage P.3 | Philippines | February, 2021 | The mutations present in this variant were the E484K and N501Y. |
Every living organism is run by a predefined set of genes that ‘instruct’ the cells on how to build the proteins. The genetic material of the coronavirus consists of ribonucleic acid (RNA) strands. Each virus has about 26,000 to 32,000 bases or RNA “letters” in its length. These letters —A, C, U and G —stand for adenine, cytosine, uracil and guanine, which are the fundamental units of the genetic code. The arrangement of A, C, U and G in the genetic code determines what proteins are expressed by the organism. So what makes these variants differ from each other?
With respect to SARS-CoV-2, a ‘Missense mutation’ makes the variants different from each other and subsequently from the original virus. A ‘Missense mutation’ is a mutation where a change in the single base pair (A unit of DNA, with 2 nucleotides bound together by hydrogen bonds) leads to the substitution of different amino acid, causing a change in the final product.
There are 10 notable missense mutations that have occurred in SARS-CoV-2:
1. N440K
2. L452R
3. S477G/N
4. E484K
5. E484Q
6. N501Y
7. D614G
8. P681H
9. P681R
10. A701V
The alphanumeric naming pertains to the amino acids replaced and the position at which they were replaced. For example, the L452R mutation describes an exchange where the Leucine (L) is replaced by Arginine(R) at position 452.
The table below displays which mutation is in which variants.
| S.No | Name of Mutation | Variant Present In |
| 1 | N440K | B.1.617 |
| 2 | L452R | B.1.429, B.1.617, |
| 3 | S477G/N | B.1.526, |
| 4 | E484K | B.1.351, P.1, B.1.525, B.1.1.7 |
| 5 | E484Q | B.1.617 |
| 6 | N501Y | B.1.1.7, B.1.351, P.1 |
| 7 | D614G | B.1.617 |
| 8 | P681H | B.1.1.7, B.1.1.207 |
| 9 | P681R | B.1.617 |
| 10 | A701V | B.1.351, B.1.526, |
Each mutation changes how the virus functions. Some mutations make the virus more transmissible. Whereas others make it more lethal and resistant to drugs and vaccines.
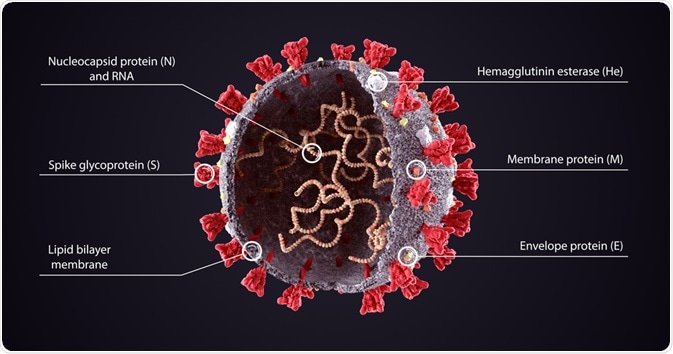
Image Credit: Orpheus FX/Shutterstock.com
So which part of the virus is being mutated? The answer to that is the spike protein. The spike protein is a linear chain of approximately 1300 amino acids folded into a structure that has 23 molecules of sugar attached to it. These polysaccharide molecules help in camouflaging the virus, and in turn evading the surveillance of the host immune system during entry. This structure gives the coronavirus the distinctive halo-like structure shown in Diagram 1.
The spike can be divided into different functional units, known as domains. There are two domains: S1 and S2. While the S1 domain serves the function of receptor-binding, the S2 domain serves the function of membrane fusion. It is observed that the receptor-binding domain (S1) is bound strongly to the ACE2 receptor (This is an enzyme receptor that acts as a lock for the spike protein of the virus which is the key. Through this the virus can infect cells) both in humans and bats. The change in the genetic code of the spike protein mutates it, thus mutating the entire virus.
All the variants are being closely monitored and daily updates are made on your country’s Ministry of Health (MoH) page. To receive up to date information, please visit the MoH site.
All we, as a community of people, can do to stop the further spread of the virus is to stay at home, respect and follow the guidelines as suggested by our respective governments and get vaccinated when the opportunity arises.
Update:
As of publishing this blog, 2 more variants have been identified. The first variant is known as the Thai variant(C.36.3). The variant has been classified as a VUI and was discovered on the 21st of May in Egypt. The people infected were tourists from Thailand.
A second variant known as the Vietnam variant was discovered on the 29th of May. Initial data suggests that it is a hybrid of the B.1.1.7 and the B.1.617.2 strains. The variant has not been completely sequenced as of writing this blog.
Will be updating this blog when more data is available.